Apr. 25, 2007 Research Highlight Biology
Translating DNA into protein
Researchers reveal an expanded role for RNA
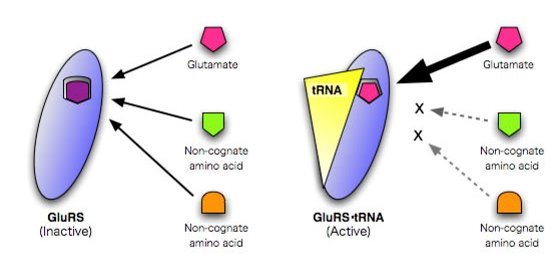 Figure 1: Specific amino acid recognition by glutamyl-tRNA synthetase depends upon it binding glutamate-tRNA.
Figure 1: Specific amino acid recognition by glutamyl-tRNA synthetase depends upon it binding glutamate-tRNA.
RIKEN researchers and Canadian colleagues have unraveled details of a mechanism for ensuring accurate translation of the genetic code into proteins. The mechanism guarantees that the molecule which transports the amino acid glutamate to a site of protein manufacture carries it and no other amino acid.
Proteins are chains constructed from 20 different kinds of amino acids. The sequence of the chain is rigidly specified by the genetic code. Each different kind of amino acid is carried by a matching transfer RNA (tRNA) molecule which fits it onto the chain. But each type of tRNA must be specific for one amino acid only. This accuracy is fundamental to protein structure and function, and hence to all cellular and bodily function.
What the research team has shown is that glutamyl-tRNA synthetase—the enzyme which controls this reaction linking glutamate to the glutamate-tRNA—first binds glutamate-tRNA. This action stimulates a change in the synthetase structure which has two consequences. It creates a pocket or binding site specific to glutamate. And it ensures the glutamate is correctly positioned to be activated by linkage with the energy compound adenosine triphosphate (ATP).
In a paper published recently in Structure 1, researchers from RIKEN’s SPring-8 Center in Harima and Genomic Sciences Center in Yokohama, and the University of Tokyo, together with colleagues from Laval University in Quebec, provided details of how they synthesized and analyzed the crystal structures of four complexes representing different combinations of the synthetase, the tRNA, glutamate and ATP. They also compared these structures with several previously described structures.
The researchers found that the complex of the synthetase and the tRNA forms a binding site which accommodates only the right form of glutamate in terms of shape, size and distribution of electrostatic charge. When not linked to the tRNA, the site can bind other incorrect amino acids (Fig. 1). So the change in structure of the synthetase upon binding the tRNA selects the right amino acid.
Once bound to the synthetase-tRNA complex, the glutamate is positioned so that it can react with ATP. In the absence of the tRNA, the ATP is bound in such a way that it can react neither with glutamate, nor any other amino acid.
A similar mechanism also seems to be in operation for the closely related amino acid glutamine and perhaps also for argenine. “It may be a basic, general mechanism,” says lead author, Shun-ichi Sekine. “I would like study the arginine complexes as a first step in checking this hypothesis.”
References
- 1. Sekine, S., Shichiri, M., Bernier, S., Chênevert, R., Lapointe, J. & Yokoyama, S. Structural bases of transfer RNA-dependent amino acid recognition and activation by glutamyl-tRNA synthetase. Structure 14, 1791–1799 (2006). doi: 10.1016/j.str.2006.10.005
