Feb. 25, 2020 Press Release Biology
New RNA mapping technique shows how RNA interacts with chromatin in the genome
A group led by scientists from the RIKEN Center for Integrative Medical Sciences (IMS) in Japan have developed a new method, RADICL-seq, which allows scientists to better understand how RNA interacts with the genome through chromatin—the structure in which the genome is organized.
There have been many changes in our understanding of the function of RNA in biology. Decades ago, it was generally believed that RNA functioned merely as an intermediary for the translation of DNA into proteins. However, the decoding of the human genome in the early 2000s led to the realization that a large amount of RNA—known as “junk” at the time—did not code for proteins. Subsequent work on large-scale genomic projects such as ENCODE and RIKEN-led FANTOM found that a large number of long non-protein coding RNAs in the mammalian genome interact with the DNA. Many of these RNAs are found in the cell nucleus, and are attached to chromatin, the structure used to tightly fold the DNA. However, it is not clear yet exactly what RNAs interact with which regions of the genomic DNA in different cells.
To achieve a better understanding of these interactions, and to determine whether RNA is actually a part of the chromatin structure, the scientists developed a new technology, which they called RNA And DNA Interacting Complexes Ligated and sequenced (RADICL-seq), which maps genome-wide RNA-chromatin interactions in intact nuclei. This technology, published in Nature Communications, allows researchers to identify distinct patterns of genome occupancy for different classes of transcripts as well as cell type-specific RNA-chromatin interactions, and emphasizes the role of transcription in the establishment of chromatin structure.
To test the validity of the method, the scientists looked at transcripts which are known to be expressed preferentially in certain cell types. The first, known at NEAT1, may be involved in the formation of a mysterious structure known as paraspeckles found in mammalian cell nuclei. The second, Fgfr2, is involved in embryonic development and tissue repair, especially for bone and blood vessels. They found that in mouse embryonic stem cells—an early type of cell—that NEAT1 acts almost exclusively on genomic regions of chromosome 19, from which it itself derives from, whereas in oligodendrocyte progenitor cells—a later type of developmental cell that can differentiate into brain cells—it interacts with a broad range of genomic regions on other chromosomes as well. Fgfr2, by contrast, mostly interacts with genomic regions on its own chromosome.
“This study is a first step toward understanding how the interplay between RNA and chromatin ensures proper genome function. Our data indicate that RNAs may exert more widespread effects on gene regulation and chromatin organization than previously thought,” says Alessandro Bonetti, one of the corresponding authors of the study.
“The broad, genome-wide applications of this technology will help us to understand the fundamental role of non-coding RNA as a regulator of genome activity, which could lead to future applications and therapies” says Piero Carninci, one of the senior authors of the study.
The work was done by scientists from RIKEN IMS along with scientists from Karolinska Institutet (Sweden), McGill University (Canada), Fondazione Santa Lucia (Italy), KAUST (Saudi Arabia), The Francis Crick Institute (United Kingdom), Imperial College (United Kingdom) and the Institute of Bioengeneering (Russia). The authors have published datasets related to the work, which can be reanalyzed by the community and are available in the GEO database (https://www.ncbi.nlm.nih.gov/geo/) using the access number GSE132192.
Reference
- Bonetti et al. (2020) RADICL-seq identifies general and cell type–specific principles of genome-wide RNA-chromatin interactions. Nat Commun. doi: 10.1038/s41467-020-14337-6
Contact
Team Leader
Piero Carninci
Laboratory for Transcriptome Technology
Center for Integrative Medical Sciences
Visiting Scientist
Alessandro Bonetti
Laboratory for Transcriptome Technology
Center for Integrative Medical Sciences
Jens Wilkinson
RIKEN International Affairs Division
E-mail: gro-pr[at]riken.jp
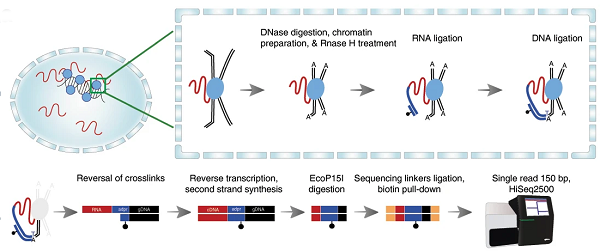
Overview of RADICL-seq technology. Top left: crosslinking of RNA bounds to DNA-binding proteins in the nucleus. Top right: a schematic explanation of the reactions that happen in crosslinked nuclei, where RNA (red) is immobilized to proteins (blue) that in turn bound the genomics DNA that constitutes the chromatin. RNAseH is a specific step introduced in RADICL-seq to reduce the amount of nascent RNA (RNA simply being transcribed everywhere in the genome) in the libraries; DNAse introduced cuts in DNA, making it available for further ligations. A linker (in blue color) ligates specifically RNA at one site and DNA at the other site: this is the key process to isolate the RNA that was bound to the chromatin. Bottom part: after removal of the proteins by “decrosslinking”, a series of reactions make the RNA (now transformed into cDNA) – DNA chimeric molecule available for deep sequencing with next generation sequencing (bottom-right). Usually 200 million reads identifies a large number of interactions in each cell type.
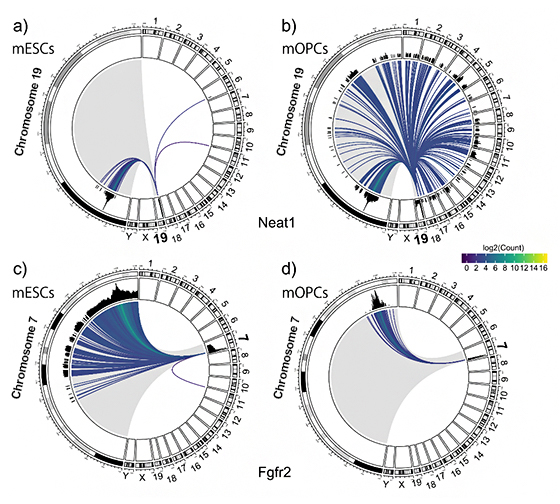
Circos plots depicting RNA-chromatin interactions mediated by Neat1 (top) and Fgfr2 (bottom) in mouse embryonic stem cells (mESCs) and oligodendrocyte progenitor cells (mOPCs), respectively. In particular, the plots show Neat1 produced from chromosome 19, interacting with a somehow narrow region of chromosome 19 and only two more loci in the Chromosome 6 and 11 in mESCs; On the contrarly, in mOPCs there are many more interaction borth with the chromosome 19 and all the other chromosomes, with a massive expansion of number of interactions called “in trans” in mOPCs. In the bottom frame, the Fgfr2 interacts mostly with loci on the same chromosome of origin (Chr. 7), with different patterns in different cells. Patterns like this can be produced for a large number of RNA-chromatin loci interactions.
