Jul. 28, 2017 Research Highlight Biology Computing / Math
Building a virtual organelle
Computer simulations expose the mechanisms underlying the disassembly and reassembly of a complex cellular organelle
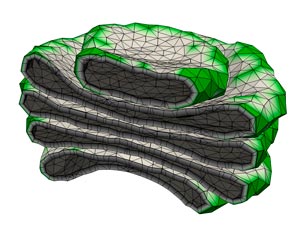 Figure 1: A computer simulation of the Golgi apparatus allowed researchers to recreate the assembly process for this complex organelle. © 2017 RIKEN Theoretical Biology Laboratory
Figure 1: A computer simulation of the Golgi apparatus allowed researchers to recreate the assembly process for this complex organelle. © 2017 RIKEN Theoretical Biology Laboratory
Every time a mammalian cell divides, it must deconstruct and then rebuild the complex protein-processing machinery of the Golgi apparatus. Computational simulations have allowed two RIKEN scientists to identify critical factors that govern this process1.
The Golgi consists of a stack of interconnected membrane-bound structures, which act as an assembly line that uses enzymes to convert newly made proteins into their fully functional, mature forms. Scientists have struggled to understand both how this organelle breaks down during cell division and how it re-forms in the resulting daughter cells.
Masashi Tachikawa, a researcher in Atsushi Mochizuki’s Theoretical Biology Laboratory, found this a daunting question to tackle. “It seemed too complicated to simulate,” he recalls.
But the researchers learned of experimental findings suggesting that this process is primarily self-governed by physical forces, rather than actively stage-managed by the cell. This meant the process could realistically be modeled if one could identify the essential ‘rules’.
Tachikawa developed a simulation that accounted for the physical and thermodynamic features of the Golgi membrane, as well as the effects of proteins that bind to and reshape the membrane. The calculations were computationally demanding, requiring a week to complete a full simulation, but they were ultimately successful.
 Figure 2: Masashi Tachikawa, RIKEN Theoretical Biology Laboratory © 2017 RIKEN
Figure 2: Masashi Tachikawa, RIKEN Theoretical Biology Laboratory © 2017 RIKEN
“We first constructed a physical model of the Golgi based on its stationary state (Fig. 1), before cell division,” says Tachikawa. “This model also reproduced the highly dynamic reassembly process of the mitotic Golgi—no additional assumptions were necessary.”
Through their simulation, the pair could home in on physical factors that determine whether the fragmented vesicles of the disassembled Golgi form a properly organized structure or give rise to warped and distorted structures.
Their results provide strong additional support for the self-assembly model of Golgi organization. They also demonstrate the power of using models to recreate dynamic biological processes that occur at a scale too tiny to observe with standard microscopes.
“Our simulation visualized membrane dynamics based on physical laws, helping us to image the details of the process and test hypothetical mechanisms,” says Tachikawa. He and Mochizuki are now collaborating with cell biologists to conduct experiments that could validate their computational model.
Tachikawa also hopes to extend such modeling approaches to even more complex cellular systems, through collaborations with RIKEN’s Interdisciplinary Theoretical Science and Interdisciplinary Theoretical and Mathematical Science programs. “We think there are lots of possibilities of updating cell biology using physical, mathematical and theoretical sciences,” he says.
Related contents
- Unraveling the complexity of cellular reactions
- Seeing the big picture in cells
- Scaling the Universe
References
- 1. Tachikawa, M. & Mochizuki, A. Golgi apparatus self-organizes into the characteristic shape via postmitotic reassembly dynamics. Proceedings of the National Academy of Sciences USA 114, 5177–5182 (2017). doi: 10.1073/pnas.1619264114
