Nov. 17, 2022 Research Highlight Biology
Gene-reading enzyme razes and rebuilds DNA-winding structures in its path
During DNA transcription, an enzyme takes apart nucleosomes and then puts them back together again
RIKEN researchers have shown how a gene-reading enzyme manages to dismantle bead-like structures of DNA and reassemble them at high speed, without disrupting the integrity of genomic organization1. Their findings could help inform the development of new therapies for diseases such as cancer.
Eukaryotic cells pack an awful lot of DNA into a tiny space. One way they achieve this is through the bead-on-string-like structure of chromatin—a mixture of DNA and proteins that make up chromosomes. The ‘beads’ are bundles of DNA known as nucleosomes.
While this structure is compact, nucleosomes pose a challenge to the DNA-transcribing enzyme called RNA polymerase II (Fig. 1), which is responsible for converting genetic information stored in DNA into messenger RNA transcripts. It has to pass through nucleosomes without altering their structure. But just how it achieves this had not been clear.
Now, a team led by Shun-ichi Sekine of the RIKEN Center for Biosystems Dynamics Research (BDR) has shown that RNA polymerase II first destroys and then rebuilds nucleosomes (Fig 1). This process allows RNA polymerase II to access genomic sequences while ensuring that epigenetic details in the structures are not lost.
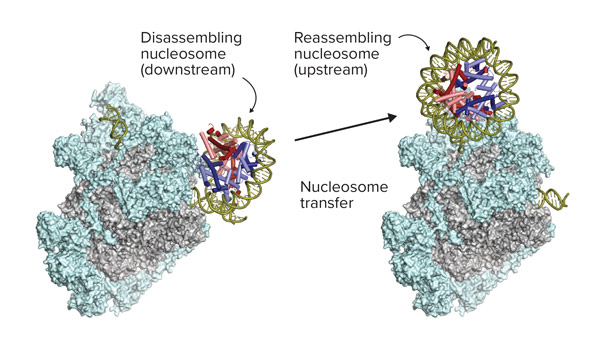
Figure 1: Structure of RNA polymerase II (gray structure) in complex with elongation factors (cyan structures), before and after passage through the nucleosome. The black arrow indicates the transcription direction. © 2022 RIKEN Center for Biosystems Dynamics Research
To discover how RNA polymerase II sidesteps the nucleosome obstacles in its path, the team turned to cryo-electron microscopy—a powerful imaging technique that can reveal the shape of biomolecules with atomic resolution. They obtained snapshots of the polymerase’s structure at six time points as it traversed the nucleosome.
The before, during and after images revealed how the polymerase, with help from several proteins known as elongation factors, first broke down a nucleosome. This allowed it to read DNA sequences that are otherwise too tightly wound up inside the nucleosomes to be accessible to the enzyme. Following transcription, the polymerase and its partner factors then reconstructed the nucleosome, aided by an extra protein called FACT, which has a role in safeguarding chromatin architecture.
“Our study reveals for the first time that RNA polymerase II disassembles the nucleosome in front of it and re-assembles it behind it, with the help of multiple other factors,” says Sekine. “As a result, the enzyme can pass through the nucleosome and continue transcription as if nothing had happened.”
The findings will be helpful for gaining new insights into diseases such as cancer that involve the improper regulation of gene transcription due to defects in nucleosome remodeling.
“The knowledge we obtained in this study could help advance research on the mechanisms of disease and aging caused by disruption of transcriptional regulation and chromatin structure,” says Haruhiko Ehara, also at BDR.
Related contents
- Elongation factors smooth transcription in the nucleosome
- Analyzing chromatin structure in more detail than ever before
- How chromosomes change their shape during cell differentiation
Rate this article
Reference
- 1. Ehara, H., Kujirai, T., Shirouzu, M., Kurumizaka, H. & Sekine, S. Structural basis of nucleosome disassembly and reassembly by RNAPII elongation complex with FACT. Science 377, eabp9466 (2022). doi: 10.1126/science.abp9466
