Jan. 27, 2022 Research Highlight Chemistry
Plausible steps toward the evolution of a key protein fold of RNA polymerases
Study sheds light on the origin of a folded structure at the heart of some key enzymes
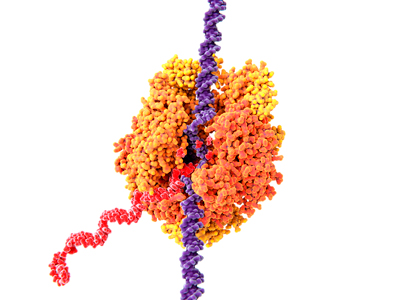 Figure 1: Illustration of RNA polymerase II (orange) in yeast. The enzyme catalyzes the transcription of DNA (purple) into messenger RNA (red). RIKEN researchers have reconstructed plausible steps for the evolutionary origin of a key protein fold in RNA polymerase. © JUAN GAERTNER/SCIENCE PHOTO LIBRARY
Figure 1: Illustration of RNA polymerase II (orange) in yeast. The enzyme catalyzes the transcription of DNA (purple) into messenger RNA (red). RIKEN researchers have reconstructed plausible steps for the evolutionary origin of a key protein fold in RNA polymerase. © JUAN GAERTNER/SCIENCE PHOTO LIBRARY
A folded region lying deep inside enzymes that are central to all forms of life could have formed more easily than previously thought, biologists at RIKEN have found1.
RNA polymerase is an enzyme that catalyzes the copying of the genetic instructions stored in DNA into working copies of the related messenger RNA (Fig. 1). Each RNA polymerase consists of multiple polypeptides and is made up of more than 3,000 amino acids.
Shunsuke Tagami of the RIKEN Center for Biosystems Dynamics Research has long been interested in the origin of RNA polymerases. “I always used to wonder how such a gigantic enzyme emerged on ancient Earth,” he recalls.
Tagami began tackling that mystery by exploring plausible evolutionary steps toward the key central region of RNA polymerase. This region is known as a double-ψ β-barrel (DPBB) as it has two neighboring symmetrical regions in which the protein chain is folded into an interlaced barrel-like structure. DPBB is found in several key proteins, suggesting that it evolved early in the origin of life.
 Sota Yagi (left), Shunsuke Tagami (right) and co-workers have reconstructed plausible steps for the evolutionary origin of a key protein fold found in RNA polymerase enzymes and several other enzymes. © 2022 RIKEN
Sota Yagi (left), Shunsuke Tagami (right) and co-workers have reconstructed plausible steps for the evolutionary origin of a key protein fold found in RNA polymerase enzymes and several other enzymes. © 2022 RIKEN
Now, Tagami and four RIKEN colleagues, along with collaborators in France, have discovered that the DPBB fold can be built from just seven of the 20 amino acids found in modern proteins. They also found that these amino acids can be coded for by only a small subset of the molecular coding features of the modern genetic code. This makes it easier to understand how this crucial protein fold could have arisen in the earliest stages of evolution.
The team’s experiments demonstrated that DPBBs can arise from the combination of two identical small sections of protein, which suggests that the ancient folded structure originated as a homodimer of a short unstructured peptide.
The researchers were surprised at how readily their reconstruction experiments succeeded. This meets the need for the earliest protein folds to have been able to form without the assistance of the complex molecular machinery of modern life. The required sections of protein also have an interesting ability to find each other and therefore ‘autopurify’ among a messy mixture of other forms.
“Our work indicates that folded proteins could have emerged much more easily than protein scientists had previously imagined,” Tagami says.
His team is now tackling the further evolutionary steps toward the much larger and more versatile and subtly regulated proteins of modern life. In the process, the team may also learn tricks to build new artificial proteins with potential uses in research, industry and medicine.
Related contents
- Polypeptide self-assembly directed by the addition of cosolvents
- Scientists identify the molecules responsible for transcriptional bursting
- A short segment of a prion protein plays a critical role in its susceptibility to cross-species prion transmission
Reference
- 1. Yagi, S., Padhi, A. K., Vucinic, J., Barbe, S., Schiex, T., Nakagawa, R., Simoncini, D., Zhang, K. Y. J. & Tagami, S. Seven amino acid types suffice to create the core fold of RNA polymerase. Journal of the American Chemical Society 143 15998–16006 (2021). doi: 10.1021/jacs.1c05367
